Comparison of data and model¶
In this tutorial, the photometric fluxes of the brown dwarf companion PZ Tel B is compared with a spectrum from the AMES-Cond atmospheric model.
Initiating species¶
[1]:
import species
[2]:
species.SpeciesInit()
Initiating species v0.1.4... [DONE]
Creating species_config.ini... [DONE]
Database: /Users/tomasstolker/applications/species/docs/tutorials/species_database.hdf5
Data folder: /Users/tomasstolker/applications/species/docs/tutorials/data
Working folder: /Users/tomasstolker/applications/species/docs/tutorials
Creating species_database.hdf5... [DONE]
Creating data folder... [DONE]
[2]:
<species.core.setup.SpeciesInit at 0x12d677438>
Adding model spectra¶
A Database object is now created which can be used for importing of data into the database.
[3]:
database = species.Database()
The spectra of the AMES-Cond model are downloaded and added to the database. This requires sufficient disk storage in the data_folder that is set in the configuration file.
While the full AMES-Cond grid is downloaded, a restiction on the wavelength and Teff range can be used when the data is imported. Also the spectral resolution can be set to downsample the high-resolution of the original spectra spectra.
[4]:
database.add_model(model='ames-cond',
wavel_range=(0.1, 6.),
spec_res=1000.,
teff_range=(2800., 3000.))
Downloading AMES-Cond model spectra (823 MB)... [DONE]
Unpacking AMES-Cond model spectra (823 MB)... [DONE]
Adding AMES-Cond model spectra... [DONE]
Adding companion data¶
The distance and magnitudes of PZ Tel B are added to the database. This will automatically download the filter transmission profiles and a flux calibrated spectrum of Vega, which are used for the flux calibration.
[5]:
database.add_companion(name='PZ Tel B')
Adding filter: Paranal/SPHERE.ZIMPOL_R_PRIM... [DONE]
Downloading Vega spectrum (270 kB)... [DONE]
Adding Vega spectrum... [DONE]
Adding filter: Paranal/SPHERE.ZIMPOL_I_PRIM... [DONE]
Adding filter: Paranal/SPHERE.IRDIS_D_H23_2... [DONE]
Adding filter: Paranal/SPHERE.IRDIS_D_H23_3... [DONE]
Adding filter: Paranal/SPHERE.IRDIS_D_K12_1... [DONE]
Adding filter: Paranal/SPHERE.IRDIS_D_K12_2... [DONE]
Adding filter: Paranal/NACO.J... [DONE]
Adding filter: Paranal/NACO.H... [DONE]
Adding filter: Paranal/NACO.Ks... [DONE]
Adding filter: Paranal/NACO.Lp... [DONE]
Adding filter: Paranal/NACO.NB405... [DONE]
Adding filter: Paranal/NACO.Mp... [DONE]
Adding filter: Gemini/NICI.ED286... [DONE]
Adding filter: Gemini/NIRI.H2S1v2-1-G0220... [DONE]
Adding object: PZ Tel B... [DONE]
Alternatively, the add_object method of Database can be used for manually adding magnitudes and spectra of an individual object. Before coninuing, let’s check the content of the database.
[6]:
database.list_content()
Database content:
- filters: <HDF5 group "/filters" (2 members)>
- Gemini: <HDF5 group "/filters/Gemini" (2 members)>
- NICI.ED286: <HDF5 dataset "NICI.ED286": shape (2, 387), type "<f8">
- NIRI.H2S1v2-1-G0220: <HDF5 dataset "NIRI.H2S1v2-1-G0220": shape (2, 129), type "<f8">
- Paranal: <HDF5 group "/filters/Paranal" (12 members)>
- NACO.H: <HDF5 dataset "NACO.H": shape (2, 23), type "<f8">
- NACO.J: <HDF5 dataset "NACO.J": shape (2, 20), type "<f8">
- NACO.Ks: <HDF5 dataset "NACO.Ks": shape (2, 27), type "<f8">
- NACO.Lp: <HDF5 dataset "NACO.Lp": shape (2, 31), type "<f8">
- NACO.Mp: <HDF5 dataset "NACO.Mp": shape (2, 18), type "<f8">
- NACO.NB405: <HDF5 dataset "NACO.NB405": shape (2, 67), type "<f8">
- SPHERE.IRDIS_D_H23_2: <HDF5 dataset "SPHERE.IRDIS_D_H23_2": shape (2, 113), type "<f8">
- SPHERE.IRDIS_D_H23_3: <HDF5 dataset "SPHERE.IRDIS_D_H23_3": shape (2, 180), type "<f8">
- SPHERE.IRDIS_D_K12_1: <HDF5 dataset "SPHERE.IRDIS_D_K12_1": shape (2, 175), type "<f8">
- SPHERE.IRDIS_D_K12_2: <HDF5 dataset "SPHERE.IRDIS_D_K12_2": shape (2, 191), type "<f8">
- SPHERE.ZIMPOL_I_PRIM: <HDF5 dataset "SPHERE.ZIMPOL_I_PRIM": shape (2, 189), type "<f8">
- SPHERE.ZIMPOL_R_PRIM: <HDF5 dataset "SPHERE.ZIMPOL_R_PRIM": shape (2, 169), type "<f8">
- models: <HDF5 group "/models" (1 members)>
- ames-cond: <HDF5 group "/models/ames-cond" (4 members)>
- flux: <HDF5 dataset "flux": shape (3, 13, 4097), type "<f8">
- logg: <HDF5 dataset "logg": shape (13,), type "<f8">
- teff: <HDF5 dataset "teff": shape (3,), type "<f8">
- wavelength: <HDF5 dataset "wavelength": shape (4097,), type "<f8">
- objects: <HDF5 group "/objects" (1 members)>
- PZ Tel B: <HDF5 group "/objects/PZ Tel B" (3 members)>
- Gemini: <HDF5 group "/objects/PZ Tel B/Gemini" (2 members)>
- NICI.ED286: <HDF5 dataset "NICI.ED286": shape (4,), type "<f8">
- NIRI.H2S1v2-1-G0220: <HDF5 dataset "NIRI.H2S1v2-1-G0220": shape (4,), type "<f8">
- Paranal: <HDF5 group "/objects/PZ Tel B/Paranal" (12 members)>
- NACO.H: <HDF5 dataset "NACO.H": shape (4,), type "<f8">
- NACO.J: <HDF5 dataset "NACO.J": shape (4,), type "<f8">
- NACO.Ks: <HDF5 dataset "NACO.Ks": shape (4,), type "<f8">
- NACO.Lp: <HDF5 dataset "NACO.Lp": shape (4,), type "<f8">
- NACO.Mp: <HDF5 dataset "NACO.Mp": shape (4,), type "<f8">
- NACO.NB405: <HDF5 dataset "NACO.NB405": shape (4,), type "<f8">
- SPHERE.IRDIS_D_H23_2: <HDF5 dataset "SPHERE.IRDIS_D_H23_2": shape (4,), type "<f8">
- SPHERE.IRDIS_D_H23_3: <HDF5 dataset "SPHERE.IRDIS_D_H23_3": shape (4,), type "<f8">
- SPHERE.IRDIS_D_K12_1: <HDF5 dataset "SPHERE.IRDIS_D_K12_1": shape (4,), type "<f8">
- SPHERE.IRDIS_D_K12_2: <HDF5 dataset "SPHERE.IRDIS_D_K12_2": shape (4,), type "<f8">
- SPHERE.ZIMPOL_I_PRIM: <HDF5 dataset "SPHERE.ZIMPOL_I_PRIM": shape (4,), type "<f8">
- SPHERE.ZIMPOL_R_PRIM: <HDF5 dataset "SPHERE.ZIMPOL_R_PRIM": shape (4,), type "<f8">
- distance: <HDF5 dataset "distance": shape (2,), type "<f8">
- spectra: <HDF5 group "/spectra" (1 members)>
- calibration: <HDF5 group "/spectra/calibration" (1 members)>
- vega: <HDF5 dataset "vega": shape (3, 8827), type "<f8">
Reading model spectra¶
Model spectra are read from the database by creating an object of ReadModel. The model name and optionally a wavelength range should be provided. The wavelength range should fit within the range that was specified in add_model.
[7]:
readmodel = species.ReadModel(model='ames-cond',
wavel_range=(0.1, 6.))
Before extracting a spectrum, let’s check which parameters are required for the AMES-Cond models.
[8]:
readmodel.get_parameters()
[8]:
['teff', 'logg']
And also the grid boundaries that are stored in the database.
[9]:
readmodel.get_bounds()
[9]:
{'teff': (2800.0, 3000.0), 'logg': (0.0, 6.0)}
The parameters are provided in a dictionary. Make sure to chose values within the grid boundaries. The radius (RJup) and distance (pc) will scale the emitted spectrum to the observer. Without these values, the spectrum is provided at the outermost layer of the atmosphere.
[10]:
model_param = {'teff': 2900., 'logg': 4.5, 'radius': 2.2, 'distance': 47.13}
The grid of spectra is linearly interpolated at a spectral resolution of 100 and stored in a ModelBox.
[11]:
modelbox = readmodel.get_model(model_param=model_param,
spec_res=100.)
Reading companion data¶
The photometry data of PZ Tel B are also read from the database and stored in an ObjectBox.
[12]:
objectbox = database.get_object(object_name='PZ Tel B')
Getting object: PZ Tel B... [DONE]
Synthetic photometry for all filters¶
For comparison, we create synthetic photometry from the model spectrum for all filters of PZ Tel B, which are stored in a SynphotBox.
[13]:
synphotbox = species.multi_photometry(datatype='model',
spectrum='ames-cond',
filters=objectbox.filters,
parameters=model_param)
Calculating synthetic photometry... [DONE]
Creating flux residuals¶
The get_residuals function is used to calculate the difference between the observed fluxes and the synthetic photometry of the model spectrum. The residuals are stored in a ResidualsBox.
[14]:
residualsbox = species.get_residuals(datatype='model',
spectrum='ames-cond',
parameters=model_param,
filters=objectbox.filters,
objectbox=objectbox,
inc_phot=True,
inc_spec=False)
Calculating synthetic photometry... [DONE]
Calculating residuals... [DONE]
Residuals [sigma]:
- Gemini/NICI.ED286: 1.55
- Gemini/NIRI.H2S1v2-1-G0220: 0.03
- Paranal/NACO.H: -0.07
- Paranal/NACO.J: -0.52
- Paranal/NACO.Ks: 1.54
- Paranal/NACO.Lp: 0.11
- Paranal/NACO.Mp: 6.60
- Paranal/NACO.NB405: 0.86
- Paranal/SPHERE.IRDIS_D_H23_2: 0.74
- Paranal/SPHERE.IRDIS_D_H23_3: 0.21
- Paranal/SPHERE.IRDIS_D_K12_1: 1.56
- Paranal/SPHERE.IRDIS_D_K12_2: 1.10
- Paranal/SPHERE.ZIMPOL_I_PRIM: -12.56
- Paranal/SPHERE.ZIMPOL_R_PRIM: -7.94
Opening Box objects¶
The open_box function can be used to view the content of a Box object. For example, the ModelBox contains several attributes, including the wavelength points and flux densities.
[15]:
modelbox.open_box()
Opening ModelBox...
model = ames-cond
type = None
wavelength = [0.10201 0.1030301 0.1040604 0.10510101 0.10615202 0.10721354
0.10828567 0.10936853 0.11046221 0.11156683 0.1126825 0.11380933
0.11494742 0.1160969 0.11725786 0.11843044 0.11961475 0.1208109
0.122019 0.12323919 0.12447159 0.1257163 0.12697346 0.1282432
0.12952563 0.13082089 0.1321291 0.13345039 0.13478489 0.13613274
0.13749407 0.13886901 0.1402577 0.14166028 0.14307688 0.14450765
0.14595272 0.14741225 0.14888637 0.15037524 0.15187899 0.15339778
0.15493176 0.15648107 0.15804589 0.15962634 0.16122261 0.16283483
0.16446318 0.16610781 0.16776889 0.16944658 0.17114105 0.17285246
0.17458098 0.17632679 0.17809006 0.17987096 0.18166967 0.18348637
0.18532123 0.18717444 0.18904619 0.19093665 0.19284602 0.19477448
0.19672222 0.19868944 0.20067634 0.2026831 0.20470993 0.20675703
0.2088246 0.21091285 0.21302198 0.2151522 0.21730372 0.21947675
0.22167152 0.22388824 0.22612712 0.22838839 0.23067227 0.232979
0.23530879 0.23766187 0.24003849 0.24243888 0.24486327 0.2473119
0.24978502 0.25228287 0.2548057 0.25735376 0.25992729 0.26252657
0.26515183 0.26780335 0.27048138 0.2731862 0.27591806 0.27867724
0.28146401 0.28427865 0.28712144 0.28999265 0.29289258 0.29582151
0.29877972 0.30176752 0.30478519 0.30783304 0.31091137 0.31402049
0.31716069 0.3203323 0.32353562 0.32677098 0.33003869 0.33333908
0.33667247 0.34003919 0.34343958 0.34687398 0.35034272 0.35384615
0.35738461 0.36095845 0.36456804 0.36821372 0.37189586 0.37561481
0.37937096 0.38316467 0.38699632 0.39086628 0.39477495 0.39872269
0.40270992 0.40673702 0.41080439 0.41491244 0.41906156 0.42325217
0.4274847 0.43175954 0.43607714 0.44043791 0.44484229 0.44929071
0.45378362 0.45832146 0.46290467 0.46753372 0.47220905 0.47693114
0.48170046 0.48651746 0.49138264 0.49629646 0.50125943 0.50627202
0.51133474 0.51644809 0.52161257 0.52682869 0.53209698 0.53741795
0.54279213 0.54822005 0.55370225 0.55923928 0.56483167 0.57047998
0.57618478 0.58194663 0.5877661 0.59364376 0.5995802 0.605576
0.61163176 0.61774808 0.62392556 0.63016481 0.63646646 0.64283113
0.64925944 0.65575203 0.66230955 0.66893265 0.67562197 0.68237819
0.68920198 0.696094 0.70305494 0.71008548 0.71718634 0.7243582
0.73160179 0.7389178 0.74630698 0.75377005 0.76130775 0.76892083
0.77661004 0.78437614 0.7922199 0.8001421 0.80814352 0.81622495
0.8243872 0.83263108 0.84095739 0.84936696 0.85786063 0.86643924
0.87510363 0.88385466 0.89269321 0.90162014 0.91063635 0.91974271
0.92894014 0.93822954 0.94761183 0.95708795 0.96665883 0.97632542
0.98608867 0.99594956 1.00590905 1.01596815 1.02612783 1.03638911
1.046753 1.05722053 1.06779273 1.07847066 1.08925537 1.10014792
1.1111494 1.12226089 1.1334835 1.14481834 1.15626652 1.16782918
1.17950748 1.19130255 1.20321558 1.21524773 1.22740021 1.23967421
1.25207095 1.26459166 1.27723758 1.29000996 1.30291006 1.31593916
1.32909855 1.34238953 1.35581343 1.36937156 1.38306528 1.39689593
1.41086489 1.42497354 1.43922327 1.45361551 1.46815166 1.48283318
1.49766151 1.51263813 1.52776451 1.54304215 1.55847257 1.5740573
1.58979787 1.60569585 1.62175281 1.63797034 1.65435004 1.67089354
1.68760248 1.7044785 1.72152329 1.73873852 1.75612591 1.77368716
1.79142404 1.80933828 1.82743166 1.84570598 1.86416304 1.88280467
1.90163271 1.92064904 1.93985553 1.95925409 1.97884663 1.99863509
2.01862144 2.03880766 2.05919573 2.07978769 2.10058557 2.12159142
2.14280734 2.16423541 2.18587777 2.20773654 2.22981391 2.25211205
2.27463317 2.2973795 2.3203533 2.34355683 2.3669924 2.39066232
2.41456894 2.43871463 2.46310178 2.4877328 2.51261013 2.53773623
2.56311359 2.58874472 2.61463217 2.64077849 2.66718628 2.69385814
2.72079672 2.74800469 2.77548474 2.80323958 2.83127198 2.8595847
2.88818055 2.91706235 2.94623298 2.97569531 3.00545226 3.03550678
3.06586185 3.09652047 3.12748567 3.15876053 3.19034813 3.22225162
3.25447413 3.28701887 3.31988906 3.35308795 3.38661883 3.42048502
3.45468987 3.48923677 3.52412914 3.55937043 3.59496413 3.63091377
3.66722291 3.70389514 3.74093409 3.77834343 3.81612687 3.85428814
3.89283102 3.93175933 3.97107692 4.01078769 4.05089557 4.09140452
4.13231857 4.17364175 4.21537817 4.25753195 4.30010727 4.34310835
4.38653943 4.43040482 4.47470887 4.51945596 4.56465052 4.61029702
4.6564 4.702964 4.74999363 4.79749357 4.84546851 4.89392319
4.94286242 4.99229105 5.04221396 5.0926361 5.14356246 5.19499808
5.24694806 5.29941755 5.35241172 5.40593584 5.4599952 5.51459515
5.5697411 5.62543851 5.6816929 5.73850982 5.79589492 5.85385387
5.91239241]
flux = [7.09673588e-05 2.11601318e-05 5.87271986e-06 2.86640705e-06
1.15608214e-06 4.04827004e-07 1.71081208e-07 4.30233634e-08
3.11527363e-08 8.06211200e-09 5.88817870e-10 2.64102520e-10
4.09799777e-10 6.55161886e-11 2.80720501e-11 1.21560648e-11
6.72024033e-12 4.60799734e-14 9.10402289e-13 6.00403269e-14
3.97723037e-19 1.04348482e-19 1.61972942e-16 2.15398100e-15
9.53305386e-15 2.66485979e-17 2.32474853e-17 5.18961551e-20
1.16259410e-20 7.47487980e-20 1.29706420e-16 2.81377619e-20
8.61317856e-21 5.32566164e-20 9.94287730e-18 4.58996127e-22
4.21361720e-23 1.49861717e-18 1.08242142e-23 6.96236018e-23
2.69982236e-24 4.03566692e-20 1.03037821e-23 7.05753437e-24
8.95449031e-24 1.56843108e-23 2.43544857e-22 2.63164650e-22
1.04809159e-22 2.79666242e-22 6.33742450e-23 2.48468930e-22
2.35894653e-22 3.84267099e-22 5.67404678e-23 1.84085447e-24
9.50194255e-24 1.47314710e-24 1.52997593e-24 5.14311531e-25
4.98782879e-25 1.82709185e-24 2.18527641e-25 2.13657960e-26
1.63342090e-26 4.42066436e-25 2.06815416e-26 3.05346806e-26
4.21992484e-26 5.50969282e-26 4.70224373e-26 1.15675678e-25
1.24523152e-24 1.83302494e-24 2.76257180e-24 2.10553262e-24
2.45519198e-23 8.08118856e-24 3.04654672e-24 9.42498387e-23
5.15058763e-24 2.15326064e-22 1.16827505e-22 5.88072075e-22
1.19060834e-22 3.13886555e-23 3.87945191e-21 1.19668998e-20
1.64651630e-20 4.56189318e-21 3.02019899e-21 1.51719418e-20
2.70952263e-20 1.39014187e-20 3.19438529e-19 1.62089074e-19
1.34600729e-19 5.20571621e-19 4.76633602e-19 3.10418608e-19
7.10317864e-19 3.88314433e-19 3.88004762e-19 2.29049034e-19
9.39887623e-19 2.98506463e-18 5.04251502e-18 3.29828069e-18
2.58007876e-18 2.63579296e-18 5.18935247e-18 5.73196151e-18
1.34414691e-17 3.48015033e-17 6.87714202e-17 5.95703984e-17
5.80816977e-17 1.54757961e-16 2.22000423e-16 3.08600639e-16
1.24119341e-16 1.73682200e-16 9.19332694e-17 9.74570974e-17
1.39767472e-16 2.96865206e-16 1.05215049e-16 1.44403290e-16
3.87703313e-16 4.67507704e-16 1.32576358e-16 2.23922826e-16
6.88855918e-16 2.95943335e-16 2.37459206e-16 3.45754334e-16
2.26599578e-16 1.25817919e-15 1.22915736e-15 1.75573081e-15
1.67672188e-15 1.55354076e-15 8.69235639e-16 1.89571686e-16
6.86336916e-16 1.38545801e-15 2.11523622e-15 1.92567722e-15
1.78099724e-15 2.53460318e-15 2.26408744e-15 2.19096659e-15
2.14954766e-15 2.14533928e-15 2.32826552e-15 2.00451765e-15
1.36463241e-15 2.84591207e-15 4.03490676e-15 2.79669131e-15
1.85964768e-15 2.37211657e-15 3.60201496e-15 2.15720311e-15
3.10141478e-15 4.10203686e-15 5.15033057e-15 6.32218545e-15
5.69890855e-15 3.81756335e-15 5.42224257e-15 4.14279714e-15
3.54838792e-15 5.77089942e-15 6.48069974e-15 6.66402240e-15
2.37830137e-15 2.88645888e-15 4.90157836e-15 8.02552299e-15
8.61546100e-15 3.87458103e-15 2.54487609e-15 4.70615014e-15
5.28813498e-15 7.59228370e-15 7.93253018e-15 9.16105313e-15
9.09717415e-15 5.45400681e-15 4.37099425e-15 5.53895340e-15
6.10966888e-15 8.80621413e-15 1.51347273e-14 5.90126067e-15
4.84432711e-15 1.03994166e-14 1.99640233e-14 3.02585772e-14
3.56660397e-14 3.84009102e-14 2.37910051e-14 1.33098520e-14
1.67848583e-14 2.15286074e-14 2.58968441e-14 3.24667138e-14
3.65195762e-14 3.78054482e-14 3.70406780e-14 3.31027509e-14
3.23185522e-14 2.56010865e-14 3.17153281e-14 3.41702709e-14
3.84174169e-14 3.72411925e-14 3.12442125e-14 3.49616647e-14
3.97078883e-14 4.07413824e-14 3.79988744e-14 3.89454096e-14
4.06915072e-14 4.17946761e-14 3.86242393e-14 4.03580052e-14
3.93516681e-14 3.27590662e-14 3.52126816e-14 3.57750684e-14
3.61814557e-14 3.76914709e-14 3.92248372e-14 4.08522810e-14
4.23976031e-14 4.22195441e-14 4.30785998e-14 4.11142771e-14
3.86205163e-14 3.88023652e-14 3.80900457e-14 3.82567775e-14
3.88571566e-14 3.90133809e-14 3.76476062e-14 3.72113099e-14
3.68934653e-14 3.75981033e-14 3.76243879e-14 3.68432193e-14
3.70026972e-14 3.66304597e-14 3.65871460e-14 3.68713537e-14
3.68780401e-14 3.51875563e-14 3.20649061e-14 2.54655507e-14
2.43009584e-14 2.45902027e-14 2.56165693e-14 2.22758723e-14
2.21916127e-14 2.07283947e-14 2.11443918e-14 2.05167105e-14
2.01078790e-14 1.98257810e-14 2.03176855e-14 2.07752104e-14
2.15683300e-14 2.12538761e-14 2.12231633e-14 2.20262100e-14
2.22338864e-14 2.24209898e-14 2.24054261e-14 2.32298745e-14
2.33208857e-14 2.35247676e-14 2.32656536e-14 2.16820716e-14
1.96997418e-14 1.81108446e-14 1.64163678e-14 1.51658079e-14
1.43217745e-14 1.33376995e-14 1.26563337e-14 1.15388387e-14
1.07703240e-14 1.13953328e-14 1.13677181e-14 1.15447871e-14
1.08757613e-14 1.09329253e-14 1.02993728e-14 1.01634336e-14
9.99591643e-15 9.85534123e-15 9.72288366e-15 9.98177661e-15
9.98843719e-15 1.01486579e-14 1.02888265e-14 1.04086309e-14
1.05638290e-14 1.04566920e-14 1.05503612e-14 1.05554935e-14
1.04432359e-14 9.46703385e-15 8.92949725e-15 8.38651574e-15
7.94890105e-15 7.20166848e-15 6.62671038e-15 6.01654055e-15
5.44638498e-15 5.00633899e-15 4.49063656e-15 4.38679075e-15
4.32951017e-15 4.07118466e-15 4.27309549e-15 4.30490655e-15
4.32272269e-15 3.93354755e-15 3.68513519e-15 3.49481662e-15
3.38282809e-15 3.25357603e-15 3.12735415e-15 3.09997362e-15
3.03775536e-15 2.82826553e-15 2.79460405e-15 2.77639393e-15
2.78651408e-15 2.60534023e-15 2.67175720e-15 2.54431241e-15
2.51886801e-15 2.50217639e-15 2.48097517e-15 2.53649057e-15
2.51999459e-15 2.43866546e-15 2.43096328e-15 2.44331100e-15
2.37045791e-15 2.36176413e-15 2.32356779e-15 2.32004058e-15
2.28666503e-15 2.25565477e-15 2.24030330e-15 2.18633033e-15
2.13729708e-15 2.11061359e-15 2.06558681e-15 2.04505330e-15
2.02301200e-15 1.98799027e-15 1.94505201e-15 1.87489362e-15
1.79711153e-15 1.68520562e-15 1.59683676e-15 1.47438916e-15
1.45657084e-15 1.38222129e-15 1.33144177e-15 1.26830784e-15
1.19402599e-15 1.12970760e-15 1.08068127e-15 9.88326505e-16
9.62055595e-16 8.89473222e-16 8.93521706e-16 8.33011317e-16
8.27360556e-16 7.84761921e-16 7.67175860e-16 7.12841091e-16
7.18639709e-16 6.88544560e-16 6.63956247e-16 6.38052721e-16
6.02536905e-16 5.80234520e-16 5.61572002e-16 5.43263485e-16
5.22440366e-16 5.06407847e-16 4.92004627e-16 4.69802444e-16
4.57975498e-16 4.41489612e-16 4.18257183e-16 4.13089640e-16
3.95458960e-16 3.87001415e-16 3.74223946e-16 3.62624292e-16
3.58030101e-16]
parameters = {'teff': 2900.0, 'logg': 4.5, 'radius': 2.2, 'distance': 47.13, 'mass': 61.7508434070773}
quantity = flux
Similarly, an ObjectBox contains a dictionary with the apparent magnitudes and a dictionary with the flux densities.
[16]:
objectbox.open_box()
Opening ObjectBox...
name = PZ Tel B
filters = ['Gemini/NICI.ED286', 'Gemini/NIRI.H2S1v2-1-G0220', 'Paranal/NACO.H', 'Paranal/NACO.J', 'Paranal/NACO.Ks', 'Paranal/NACO.Lp', 'Paranal/NACO.Mp', 'Paranal/NACO.NB405', 'Paranal/SPHERE.IRDIS_D_H23_2', 'Paranal/SPHERE.IRDIS_D_H23_3', 'Paranal/SPHERE.IRDIS_D_K12_1', 'Paranal/SPHERE.IRDIS_D_K12_2', 'Paranal/SPHERE.ZIMPOL_I_PRIM', 'Paranal/SPHERE.ZIMPOL_R_PRIM']
magnitude = {'Gemini/NICI.ED286': array([11.68, 0.14]), 'Gemini/NIRI.H2S1v2-1-G0220': array([11.39, 0.14]), 'Paranal/NACO.H': array([11.93, 0.14]), 'Paranal/NACO.J': array([12.47, 0.2 ]), 'Paranal/NACO.Ks': array([11.53, 0.07]), 'Paranal/NACO.Lp': array([11.04, 0.22]), 'Paranal/NACO.Mp': array([10.93, 0.03]), 'Paranal/NACO.NB405': array([10.94, 0.07]), 'Paranal/SPHERE.IRDIS_D_H23_2': array([11.78, 0.19]), 'Paranal/SPHERE.IRDIS_D_H23_3': array([11.65, 0.19]), 'Paranal/SPHERE.IRDIS_D_K12_1': array([11.56, 0.09]), 'Paranal/SPHERE.IRDIS_D_K12_2': array([11.29, 0.1 ]), 'Paranal/SPHERE.ZIMPOL_I_PRIM': array([15.16, 0.12]), 'Paranal/SPHERE.ZIMPOL_R_PRIM': array([17.84, 0.31])}
flux = {'Gemini/NICI.ED286': array([2.78256313e-14, 3.59792047e-15]), 'Gemini/NIRI.H2S1v2-1-G0220': array([1.05904381e-14, 1.36936891e-15]), 'Paranal/NACO.H': array([1.96875866e-14, 2.54565189e-15]), 'Paranal/NACO.J': array([3.11068448e-14, 5.76255332e-15]), 'Paranal/NACO.Ks': array([1.12416273e-14, 7.25276710e-16]), 'Paranal/NACO.Lp': array([2.02006237e-15, 4.12126891e-16]), 'Paranal/NACO.Mp': array([9.18778530e-16, 2.53900194e-17]), 'Paranal/NACO.NB405': array([1.67046283e-15, 1.07773346e-16]), 'Paranal/SPHERE.IRDIS_D_H23_2': array([2.54130005e-14, 4.46991835e-15]), 'Paranal/SPHERE.IRDIS_D_H23_3': array([2.42699565e-14, 4.26886718e-15]), 'Paranal/SPHERE.IRDIS_D_K12_1': array([1.15478090e-14, 9.58329872e-16]), 'Paranal/SPHERE.IRDIS_D_K12_2': array([1.14281211e-14, 1.05405764e-15]), 'Paranal/SPHERE.ZIMPOL_I_PRIM': array([1.08628801e-14, 1.20305572e-15]), 'Paranal/SPHERE.ZIMPOL_R_PRIM': array([1.82633135e-15, 5.28569079e-16])}
distance = [47.13 0.13]
spectrum = None
The attributes in the Box objects can be extracted for further analyis or manual plots, for example:
[17]:
modelbox.wavelength
[17]:
array([0.10201 , 0.1030301 , 0.1040604 , 0.10510101, 0.10615202,
0.10721354, 0.10828567, 0.10936853, 0.11046221, 0.11156683,
0.1126825 , 0.11380933, 0.11494742, 0.1160969 , 0.11725786,
0.11843044, 0.11961475, 0.1208109 , 0.122019 , 0.12323919,
0.12447159, 0.1257163 , 0.12697346, 0.1282432 , 0.12952563,
0.13082089, 0.1321291 , 0.13345039, 0.13478489, 0.13613274,
0.13749407, 0.13886901, 0.1402577 , 0.14166028, 0.14307688,
0.14450765, 0.14595272, 0.14741225, 0.14888637, 0.15037524,
0.15187899, 0.15339778, 0.15493176, 0.15648107, 0.15804589,
0.15962634, 0.16122261, 0.16283483, 0.16446318, 0.16610781,
0.16776889, 0.16944658, 0.17114105, 0.17285246, 0.17458098,
0.17632679, 0.17809006, 0.17987096, 0.18166967, 0.18348637,
0.18532123, 0.18717444, 0.18904619, 0.19093665, 0.19284602,
0.19477448, 0.19672222, 0.19868944, 0.20067634, 0.2026831 ,
0.20470993, 0.20675703, 0.2088246 , 0.21091285, 0.21302198,
0.2151522 , 0.21730372, 0.21947675, 0.22167152, 0.22388824,
0.22612712, 0.22838839, 0.23067227, 0.232979 , 0.23530879,
0.23766187, 0.24003849, 0.24243888, 0.24486327, 0.2473119 ,
0.24978502, 0.25228287, 0.2548057 , 0.25735376, 0.25992729,
0.26252657, 0.26515183, 0.26780335, 0.27048138, 0.2731862 ,
0.27591806, 0.27867724, 0.28146401, 0.28427865, 0.28712144,
0.28999265, 0.29289258, 0.29582151, 0.29877972, 0.30176752,
0.30478519, 0.30783304, 0.31091137, 0.31402049, 0.31716069,
0.3203323 , 0.32353562, 0.32677098, 0.33003869, 0.33333908,
0.33667247, 0.34003919, 0.34343958, 0.34687398, 0.35034272,
0.35384615, 0.35738461, 0.36095845, 0.36456804, 0.36821372,
0.37189586, 0.37561481, 0.37937096, 0.38316467, 0.38699632,
0.39086628, 0.39477495, 0.39872269, 0.40270992, 0.40673702,
0.41080439, 0.41491244, 0.41906156, 0.42325217, 0.4274847 ,
0.43175954, 0.43607714, 0.44043791, 0.44484229, 0.44929071,
0.45378362, 0.45832146, 0.46290467, 0.46753372, 0.47220905,
0.47693114, 0.48170046, 0.48651746, 0.49138264, 0.49629646,
0.50125943, 0.50627202, 0.51133474, 0.51644809, 0.52161257,
0.52682869, 0.53209698, 0.53741795, 0.54279213, 0.54822005,
0.55370225, 0.55923928, 0.56483167, 0.57047998, 0.57618478,
0.58194663, 0.5877661 , 0.59364376, 0.5995802 , 0.605576 ,
0.61163176, 0.61774808, 0.62392556, 0.63016481, 0.63646646,
0.64283113, 0.64925944, 0.65575203, 0.66230955, 0.66893265,
0.67562197, 0.68237819, 0.68920198, 0.696094 , 0.70305494,
0.71008548, 0.71718634, 0.7243582 , 0.73160179, 0.7389178 ,
0.74630698, 0.75377005, 0.76130775, 0.76892083, 0.77661004,
0.78437614, 0.7922199 , 0.8001421 , 0.80814352, 0.81622495,
0.8243872 , 0.83263108, 0.84095739, 0.84936696, 0.85786063,
0.86643924, 0.87510363, 0.88385466, 0.89269321, 0.90162014,
0.91063635, 0.91974271, 0.92894014, 0.93822954, 0.94761183,
0.95708795, 0.96665883, 0.97632542, 0.98608867, 0.99594956,
1.00590905, 1.01596815, 1.02612783, 1.03638911, 1.046753 ,
1.05722053, 1.06779273, 1.07847066, 1.08925537, 1.10014792,
1.1111494 , 1.12226089, 1.1334835 , 1.14481834, 1.15626652,
1.16782918, 1.17950748, 1.19130255, 1.20321558, 1.21524773,
1.22740021, 1.23967421, 1.25207095, 1.26459166, 1.27723758,
1.29000996, 1.30291006, 1.31593916, 1.32909855, 1.34238953,
1.35581343, 1.36937156, 1.38306528, 1.39689593, 1.41086489,
1.42497354, 1.43922327, 1.45361551, 1.46815166, 1.48283318,
1.49766151, 1.51263813, 1.52776451, 1.54304215, 1.55847257,
1.5740573 , 1.58979787, 1.60569585, 1.62175281, 1.63797034,
1.65435004, 1.67089354, 1.68760248, 1.7044785 , 1.72152329,
1.73873852, 1.75612591, 1.77368716, 1.79142404, 1.80933828,
1.82743166, 1.84570598, 1.86416304, 1.88280467, 1.90163271,
1.92064904, 1.93985553, 1.95925409, 1.97884663, 1.99863509,
2.01862144, 2.03880766, 2.05919573, 2.07978769, 2.10058557,
2.12159142, 2.14280734, 2.16423541, 2.18587777, 2.20773654,
2.22981391, 2.25211205, 2.27463317, 2.2973795 , 2.3203533 ,
2.34355683, 2.3669924 , 2.39066232, 2.41456894, 2.43871463,
2.46310178, 2.4877328 , 2.51261013, 2.53773623, 2.56311359,
2.58874472, 2.61463217, 2.64077849, 2.66718628, 2.69385814,
2.72079672, 2.74800469, 2.77548474, 2.80323958, 2.83127198,
2.8595847 , 2.88818055, 2.91706235, 2.94623298, 2.97569531,
3.00545226, 3.03550678, 3.06586185, 3.09652047, 3.12748567,
3.15876053, 3.19034813, 3.22225162, 3.25447413, 3.28701887,
3.31988906, 3.35308795, 3.38661883, 3.42048502, 3.45468987,
3.48923677, 3.52412914, 3.55937043, 3.59496413, 3.63091377,
3.66722291, 3.70389514, 3.74093409, 3.77834343, 3.81612687,
3.85428814, 3.89283102, 3.93175933, 3.97107692, 4.01078769,
4.05089557, 4.09140452, 4.13231857, 4.17364175, 4.21537817,
4.25753195, 4.30010727, 4.34310835, 4.38653943, 4.43040482,
4.47470887, 4.51945596, 4.56465052, 4.61029702, 4.6564 ,
4.702964 , 4.74999363, 4.79749357, 4.84546851, 4.89392319,
4.94286242, 4.99229105, 5.04221396, 5.0926361 , 5.14356246,
5.19499808, 5.24694806, 5.29941755, 5.35241172, 5.40593584,
5.4599952 , 5.51459515, 5.5697411 , 5.62543851, 5.6816929 ,
5.73850982, 5.79589492, 5.85385387, 5.91239241])
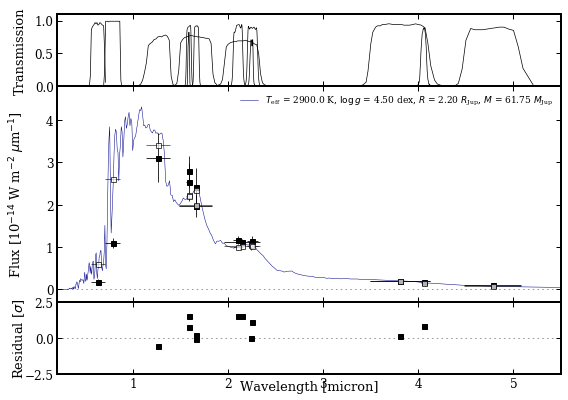
Plotting spectra, fluxes and filters¶
The Box objects can provided as list to plot_spectrum, which will be interpreted accordingly. Optionally, the filter names can be included for plotting the transmission profiles and the ResidualsBox can be included for showing the residuals. The ObjectBox requires a color for the photometry data and a spectrum with the latter set to None in this case.
[18]:
species.plot_spectrum(boxes=[modelbox, objectbox, synphotbox],
filters=objectbox.filters,
residuals=residualsbox,
colors=('darkblue', ('black', None), 'black'),
offset=(-0.08, -0.06),
xlim=(0.2, 5.5),
ylim=(-3e-15, 4.8e-14),
ylim_res=(-2.5, 2.5),
legend='upper right',
output='spectrum.png')
Plotting spectrum: spectrum.png... [DONE]
Let’s have a look at the result. The plot is stored in the working folder. The black squares are the photometry data of PZ Tel B and the open squares are the synthetic photometry computed from the model spectrum. The residuals are shown relative to the uncertainties on the fluxes.
[19]:
from IPython.display import Image
Image('spectrum.png')
[19]: