Atmospheric models¶
The DRIFT-PHOENIX model spectra are added to the database. The grid is then interpolated and a spectrum for a given set of parameter values and spectral resolution is computed. The spectrum is then plotted together with several filter profiles.
Getting started¶
[1]:
import species
from IPython.display import Image
[2]:
species.SpeciesInit()
Initiating species v0.2.0... [DONE]
Database: /Users/tomasstolker/applications/species/docs/tutorials/species_database.hdf5
Data folder: /Users/tomasstolker/applications/species/docs/tutorials/data
Working folder: /Users/tomasstolker/applications/species/docs/tutorials
[2]:
<species.core.setup.SpeciesInit at 0x124122080>
Adding model spectra to the database¶
[3]:
database = species.Database()
[4]:
database.add_model(model='drift-phoenix',
wavel_range=None,
spec_res=None,
teff_range=None)
Unpacking DRIFT-PHOENIX model spectra... [DONE]
Adding DRIFT-PHOENIX model spectra... [DONE]
Interpolating the model grid¶
[5]:
read_model = species.ReadModel(model='drift-phoenix',
wavel_range=(1., 5.))
[6]:
read_model.get_bounds()
[6]:
{'teff': (1000.0, 3000.0), 'logg': (3.0, 5.5), 'feh': (-0.6, 0.3)}
[7]:
model_param = {'teff':1510., 'logg':4.1, 'feh':0.1}
[8]:
modelbox = read_model.get_model(model_param=model_param,
spec_res=200.)
[9]:
filters = ['MKO/NSFCam.J', 'MKO/NSFCam.H', 'MKO/NSFCam.K', 'MKO/NSFCam.Lp', 'MKO/NSFCam.Mp']
[10]:
species.plot_spectrum(boxes=[modelbox, ],
filters=filters,
offset=(-0.08, -0.06),
xlim=(1., 5.),
ylim=(0., 1.1e5),
legend='upper right',
output='model_spectrum.png')
Plotting spectrum: model_spectrum.png... [DONE]
[11]:
Image('model_spectrum.png')
[11]:
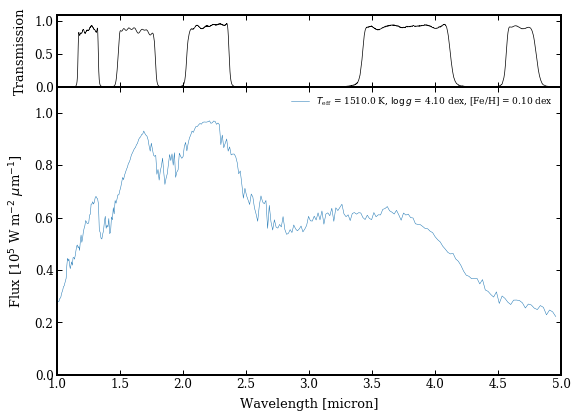
Extracting a spectrum at a grid point¶
It is also possible to extract a spectrum without interpolating at one of the grid points. Let’s first check what parameter values are available for the DRIFT-PHOENIX models.
[12]:
read_model.get_points()
[12]:
{'teff': array([1000., 1100., 1200., 1300., 1400., 1500., 1600., 1700., 1800.,
1900., 2000., 2100., 2200., 2300., 2400., 2500., 2600., 2700.,
2800., 2900., 3000.]),
'logg': array([3. , 3.5, 4. , 4.5, 5. , 5.5]),
'feh': array([-0.6, -0.3, -0. , 0.3])}
The get_data function is used for extracting an spectrum from the discrete parameter grid.
[13]:
model_param = {'teff':1500., 'logg':4., 'feh':0., 'radius':1., 'distance':20.}
[14]:
read_model = species.ReadModel(model='drift-phoenix',
wavel_range=(1., 5.))
[15]:
modelbox = read_model.get_data(model_param)
[16]:
species.plot_spectrum(boxes=[modelbox, ],
filters=filters,
offset=(-0.08, -0.06),
xlim=(1., 5.),
ylim=(0., 1.35e-15),
legend='upper right',
output='model_spectrum.png')
Plotting spectrum: model_spectrum.png... [DONE]
[17]:
Image('model_spectrum.png')
[17]:
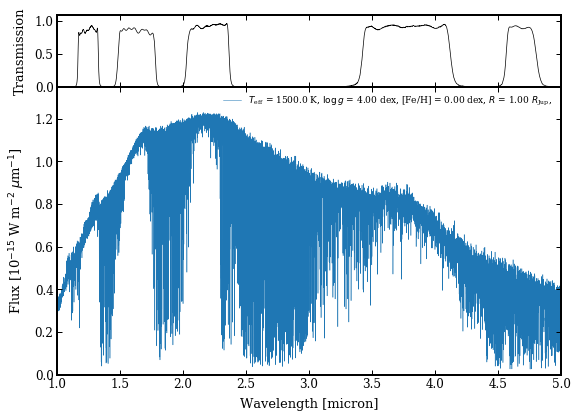
Calculating synthetic photometry¶
[18]:
read_model = species.ReadModel(model='drift-phoenix',
filter_name='Paranal/NACO.Mp')
[19]:
flux = read_model.get_flux(model_param)
print(f'Flux density [W m-2 micron-1] = {flux[0]:.2e}')
Flux density [W m-2 micron-1] = 3.43e-16
[20]:
app_mag, abs_mag = read_model.get_magnitude(model_param)
print(f'Apparent magnitude [mag] = {app_mag:.2f}')
print(f'Absolute magnitude [mag] = {abs_mag:.2f}')
Apparent magnitude [mag] = 12.00
Absolute magnitude [mag] = 10.50